Nanopore sequencing, a pivotal component of long-read sequencing technology, has emerged as a formidable and promising approach in the realm of genomics and molecular biology. Spearheaded by the Oxford Nanopore Company, this innovative technology bears the potential to revolutionize genomic research and clinical applications.
The fundamental principle of nanopore sequencing revolves around the passage of individual DNA or RNA molecules through nanometer-sized pores, enabling real-time analysis of their nucleotide sequence. In sharp contrast to first and second-generation sequencing technologies, nanopore sequencing obviates the need for PCR amplification, thereby eliminating time-consuming and potentially biased amplification steps. This direct sequencing approach empowers researchers to attain long reads, furnishing comprehensive insights into genomic regions that prove challenging to analyze using short-read sequencing methodologies.
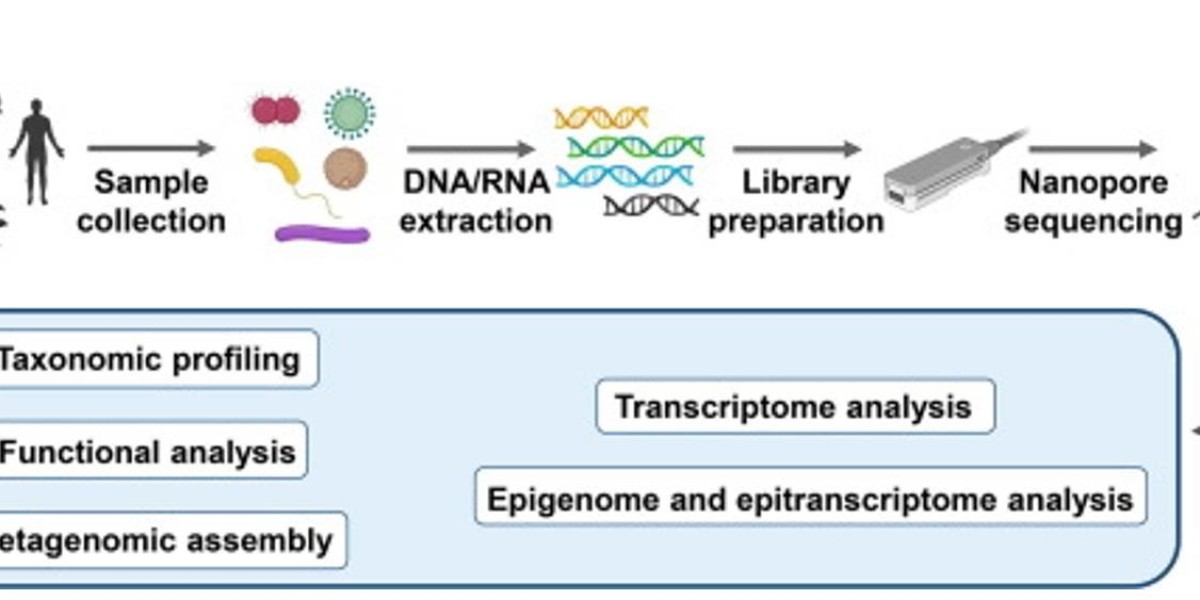
The Oxford Nanopore sequencing process and performance. (Magi et al., 2018)
Principle of Nanopore Sequencing
Nanopore sequencing represents a contemporary technique employed for DNA and RNA sequencing, predicated on the passage of nucleic acid molecules through a nanopore to discern their sequence. The process entails employing a nanopore, a minute orifice at the nanometer scale, which can either be a naturally occurring protein pore or an artificial one.
The foundational tenets of nanopore sequencing are as follows:
- Nanopore: Functioning as a single-molecule sensor, the nanopore is a diminutive aperture fabricated from diverse materials, such as protein, solid-state materials, or synthetic substances. The nanopore is equipped with a molecular junction or reader, covalently attached to the pore.
- Immobilization: In the case of protein-based nanopores, they are immobilized on a resistive membrane.
- Kinetic Proteins: Special motor proteins are employed to draw the nucleic acid (DNA or RNA) molecules through the nanopore. These motor proteins are linked to the nucleic acids, tethered to prevent them from drifting freely in the solution.
- Current Changes: As the nucleic acid transits through the nanopore, it induces a change in charge due to the inherent charge of individual bases (A, T, C, G for DNA; A, U, C, G for RNA). These charge alterations lead to modifications in the electric current flowing through the resistive membrane.
- Real-Time Monitoring: The alterations in current are continuously monitored in real-time during the nucleic acid's traversal through the nanopore.
- Base Identification: Distinctive current signals produced by different bases (A, T, C, G) facilitate the determination of the nucleic acid's sequence. Analyzing the variations in current enables the decoding of the DNA or RNA molecule's sequence.
- Sequencing: By scrutinizing these current signals, the sequence of the nucleic acid molecule can be ascertained, facilitating the sequencing of DNA or RNA.
Key components involved in nanopore sequencing include:
- Reader: Transmembrane proteins (such as protein nanopores) are used as readers to detect the nucleic acid passing through the nanopore. These reader proteins can be natural or artificial, obtained through genetic engineering modifications.
- Membrane: A synthetic polymeric membrane with high electrical resistivity is used, and the reader protein is embedded within it. An ionic solution surrounds the membrane. Applying different potentials on both sides of the membrane allows ions to flow through the nanopore, resulting in an electric current change when nucleic acids pass through.
- Motor: Motor proteins are used to attach and move DNA or RNA molecules through the nanopore during library preparation for sequencing.
- Tether: The tether is employed to anchor the DNA or RNA strand, preventing it from floating freely in the solution and guiding it into the nanopore for sequencing.
The Process of Nanopore Sequencing
The Nanopore sequencing process involves the following steps:
- DNA Denaturation: The double-stranded DNA is uncoiled and unraveled into single-stranded molecules.
- Passage through Nanopore: The single-stranded DNA molecules pass through a pore protein. This pore contains a protein molecule that acts as a converter.
- Current Modulation: As DNA molecules move through the nanopore, ions pass through as well, causing changes in the electrical current.
- Current-Based Base Identification: The different bases (A, C, G, T) of the DNA create distinct changes in the electrical current as they pass through the pore. The converter protein detects and interprets these current fluctuations for each of the five bases.
- Pattern Recognition: By analyzing the current changes, a pattern recognition algorithm is applied to deduce the base sequence of the DNA molecule.
Advantages of Nanopore Sequencing Technology
Nanopore sequencing technology boasts several distinct advantages, rendering it an exceptionally enticing approach for genetic analysis and research:
- Extended Read Length: Nanopore sequencing enables the generation of exceptionally long reads, reaching megabases in length. This extended read capability facilitates comprehensive genome analysis, allowing researchers to detect complex genomic structures and variations.
- Cost-Effectiveness: Compared to other sequencing technologies, Nanopore sequencing entails relatively low equipment costs. Notably, the sequencing chips can be cleaned and regenerated for reuse, enhancing cost-effectiveness and reducing overall operational expenses.
- Real-Time Data Access: One remarkable feature of Nanopore sequencing is the ability to obtain sequence information in real-time. This means that the sequencing process and data analysis can be completed within an impressively short timeframe, sometimes as early as one hour. Such rapid turnaround time is particularly valuable in time-critical applications where dynamic detection of large genomes is essential.
- Portability and Flexibility: Nanopore sequencing devices are compact and lightweight, enabling easy portability. This empowers researchers to conduct sequencing experiments in diverse settings and facilitates field research applications, especially in scenarios where access to fully equipped laboratories might be limited.
- Direct Sequencing of DNA and RNA: An important advantage of Nanopore sequencing is its ability to directly sequence original DNA and RNA molecules without the need for PCR amplification. This eliminates amplification biases and ensures a more accurate representation of the genetic content. Additionally, Nanopore sequencing retains the original base modification information, enabling the direct detection of methylated cytosine residues, which is critical for epigenetic studies.
What Is the Future of Nanopore Sequencing Technology?
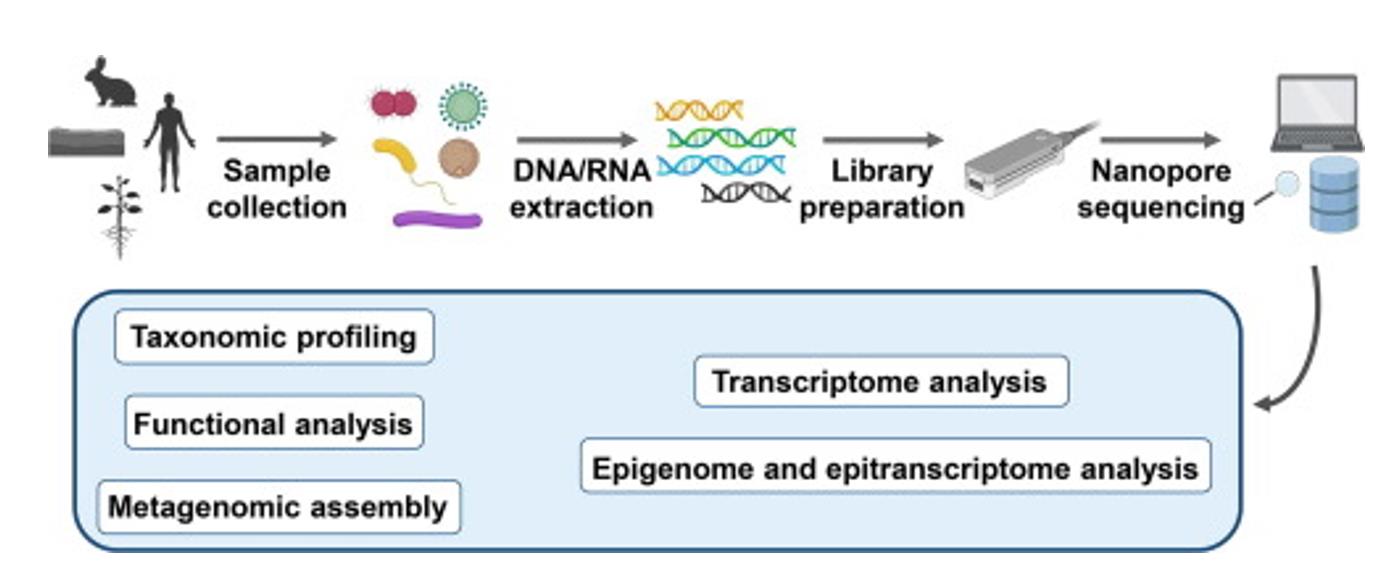
Nanopore sequencing and its application to the study of microbial communities. (Ciuffreda et al., 2021)
- Facilitating Complex Genome Splicing
Addressing the historical challenges of genome splicing, especially in polyploid, highly repetitive, and heterozygous animal and plant genomes, has been arduous with conventional short-fragment-based approaches. However, the advent of Nanopore sequencing technology has ushered in a transformative solution. Characterized by exceptionally long read lengths, Nanopore sequencing enables efficient splicing of large genomes, significantly enhancing genome integrity.
- Comprehensive Full-Length Transcriptome Analysis
Traditionally, obtaining and analyzing full-length transcripts in transcriptome analysis presented significant hurdles, largely stemming from the need to interrupt mRNA and undergo reverse transcription to cDNA. Nanopore's remarkable long read length capability now offers a straightforward and precise means to identify multiple homologous isoforms of genes. This direct sequencing of RNA not only simplifies the process but also facilitates the identification of base modifications in RNA molecules.
- Precise Detection of Large Segment Structural Variants
Human diseases often exhibit large structural variations in the genome, such as deletions, inversions, and translocations. Detecting these structural variations accurately with conventional short sequencing read lengths has remained challenging. Leveraging the longer read lengths of Nanopore sequencing, it proves highly suitable for the precise detection of these large structural variants. This promising capability holds significant potential for advancing disease research.
- Real-Time Identification of Microorganisms
Nanopore sequencing's real-time and rapid nature provides a cutting-edge approach to directly sequence genetic material at the collection point. This immediate sequence information enables swift species classification and identification of microorganisms, leading to expedited and efficient microbial identification processes.
- Single-Molecule Epigenomics
Nanopore sequencing has the potential to unlock single-molecule epigenetic analysis, providing valuable insights into DNA methylation, chromatin structure, and other epigenetic modifications. This capability could significantly impact our understanding of gene regulation and disease mechanisms.
References
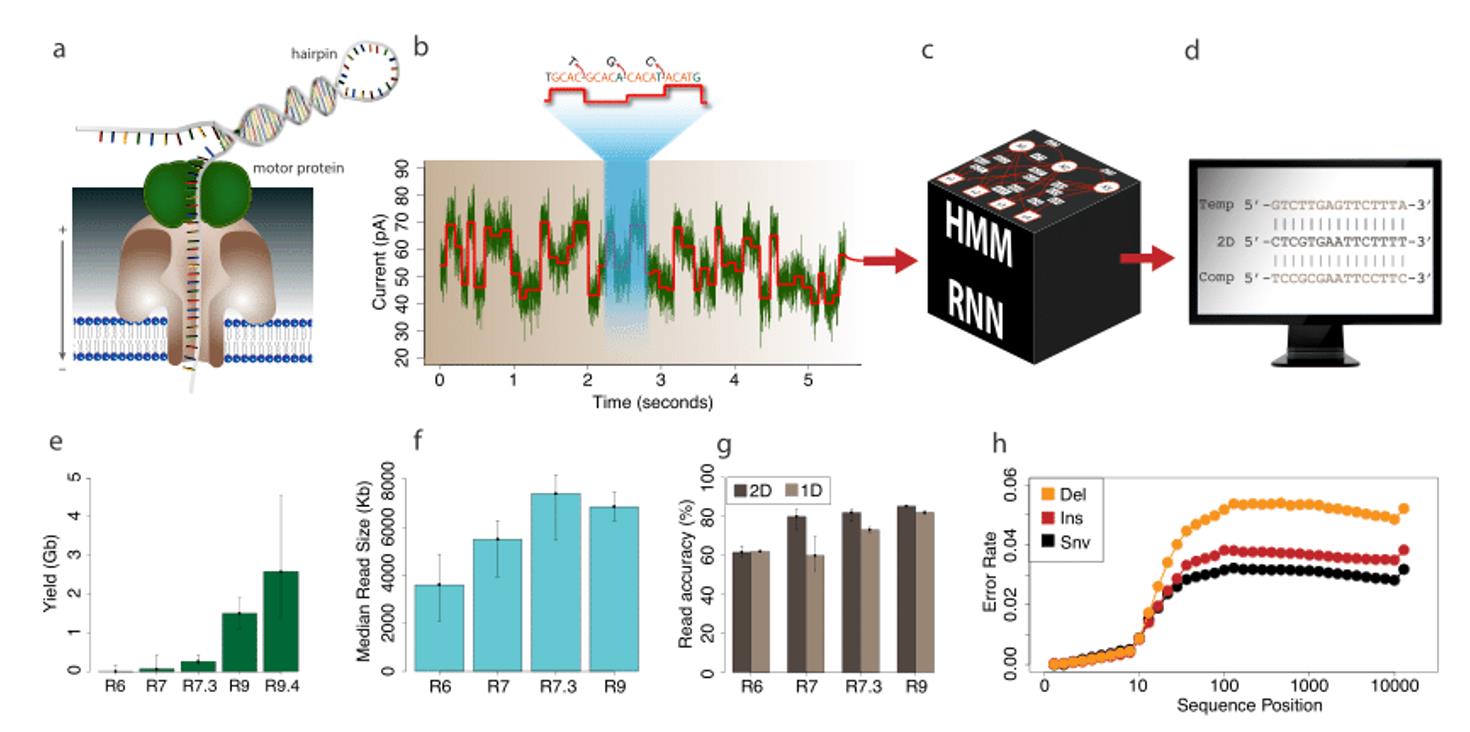
- Magi, Alberto, et al. "Nanopore sequencing data analysis: state of the art, applications and challenges." Briefings in bioinformatics 19.6 (2018): 1256-1272.
- Ciuffreda, Laura, Héctor Rodríguez-Pérez, and Carlos Flores. "Nanopore sequencing and its application to the study of microbial communities."